2020-03-16
这是一个 R 语言包,使用教程详见 https://openr.pzhao.org/zh/tags/ncovr/。这里是个简介。
ncovr 包是方便 R 用户获取新型冠状病毒(2019-nCoV)数据而开发的,后续增添了数据处理、建模、可视化等功能。
数据获取途径
ncovr 包获取数据的主要途径是 BlankerL/DXY-2019-nCoV-Crawler。这个项目提供了 api 接口和 csv 文件。为了减轻 api 的流量压力, ncovr 每天将每天自动从这个 api 读一次数据,保存成 R 语言直接读取的 .RDS 格式,方便 R 语言用户调用。详见下面的示例。
安装
安装 R。在 CRAN 上选择适合你操作系统的安装包来安装。
安装 remotes 包:
install.packages('remotes')安装 ncovr 包:
remotes::install_github('pzhaonet/ncovr')
获取数据
# Sys.setlocale('LC_CTYPE', 'Chinese') # windows <U+7528><U+6237><U+8BBE><U+7F6E><U+4E2D><U+6587><U+73AF><U+5883> require("ncovr")
require("leafletCN")
ncov <- get_ncov() # <U+8BFB><U+53D6> RDS<U+6570><U+636E>(<U+63A8><U+8350>)<U+56FD><U+5185><U+4ECE>github<U+8BFB><U+53D6><U+6570><U+636E><U+4E0D><U+7A33><U+5B9A>! # get_ncov(method = 'csv') # <U+4ECE> csv <U+6587><U+4EF6><U+8BFB><U+53D6>(<U+63A8><U+8350>) # get_ncov(method = 'api') # <U+4ECE> api <U+63A5><U+53E3><U+8BFB><U+53D6>
另外提供了下载数据的函数get_ncov2(),定义新的 ncov 类(实际也是data frame),提供了subset()方法, 用于提取 ncov 子集,参数可以是省份、china或者world
ncov2 <- get_ncov2() ncov2
## All COVID 2019 Data
## Updated at 2020-03-16 15:44:16
## From https://github.com/yiluheihei/nCoV-2019-Data# <U+6E56><U+5317> ncov, <U+6309><U+5E02><U+7EDF><U+8BA1> hubei_ncov <- ncov2["<U+6E56><U+5317>"] hubei_ncov
## Hubei COVID 2019 Data
## Updated at 2020-03-16 13:54:01
## From https://github.com/yiluheihei/nCoV-2019-Datahead(data.frame(hubei_ncov), 5)
## cityEnglishName cityName provinceName provinceShortName provinceEnglishName
## 1 Wuhan 武汉 湖北省 湖北 Hubei
## 2 Xiaogan 孝感 湖北省 湖北 Hubei
## 3 Ezhou 鄂州 湖北省 湖北 Hubei
## 4 Suizhou 随州 湖北省 湖北 Hubei
## 5 Jingzhou 荆州 湖北省 湖北 Hubei
## currentConfirmedCount confirmedCount suspectedCount curedCount deadCount
## 1 9102 50003 0 38432 2469
## 2 125 3518 0 3266 127
## 3 62 1394 0 1275 57
## 4 41 1307 0 1221 45
## 5 38 1580 0 1492 50
## updateTime
## 1 2020-03-16 13:54:01
## 2 2020-03-16 13:54:01
## 3 2020-03-16 13:54:01
## 4 2020-03-16 13:54:01
## 5 2020-03-16 13:54:01# china,<U+6309><U+7701><U+7EDF><U+8BA1> china_ncov <- ncov2["china"] head(data.frame(china_ncov), 5)
## provinceName provinceShortName provinceEnglishName currentConfirmedCount
## 1 中国 中国 China 9951
## 2 湖北省 湖北 Hubei 9557
## 3 辽宁省 辽宁 Liaoning 9
## 4 广西壮族自治区 广西 Guangxi 2
## 5 海南省 海南 Hainan 1
## confirmedCount suspectedCount curedCount deadCount updateTime
## 1 81099 0 67930 3218 2020-03-16 13:54:38
## 2 67798 0 55142 3099 2020-03-16 13:54:01
## 3 125 0 115 1 2020-03-16 13:54:01
## 4 252 0 248 2 2020-03-16 13:49:01
## 5 168 0 161 6 2020-03-16 13:49:01# world, <U+6309><U+56FD><U+5BB6><U+7EDF><U+8BA1> world_ncov <- ncov2["world"]
## Parsed with column specification:
## cols(
## countryName = col_character(),
## countryEnglishName = col_character()
## )head(data.frame(world_ncov), 5)
## countryEnglishName countryName currentConfirmedCount confirmedCount
## 1 Afghanistan 阿富汗 16 16
## 2 Albania 阿尔巴尼亚 41 42
## 3 Algeria 阿尔及利亚 41 54
## 4 Andorra 安道尔 1 2
## 5 Antigua and Barbuda 安提瓜和巴布达 1 1
## suspectedCount curedCount deadCount updateTime
## 1 0 0 0 2020-03-16 06:53:45
## 2 0 0 1 2020-03-16 06:53:45
## 3 0 10 3 2020-03-16 06:53:45
## 4 0 1 0 2020-03-16 06:53:45
## 5 0 0 0 2020-03-16 06:53:45国家地图
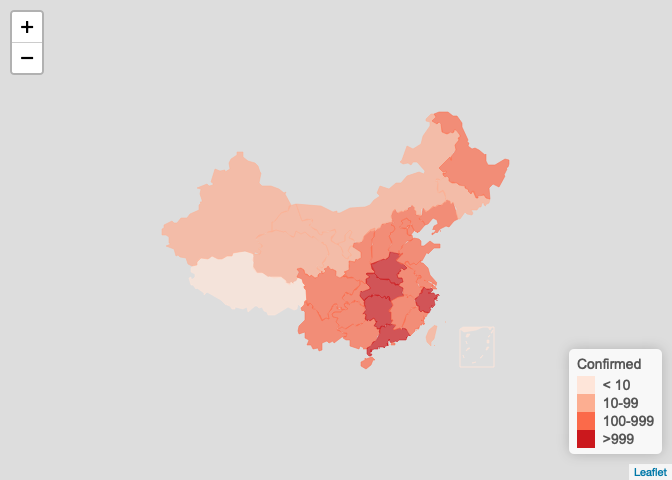
按省级显示
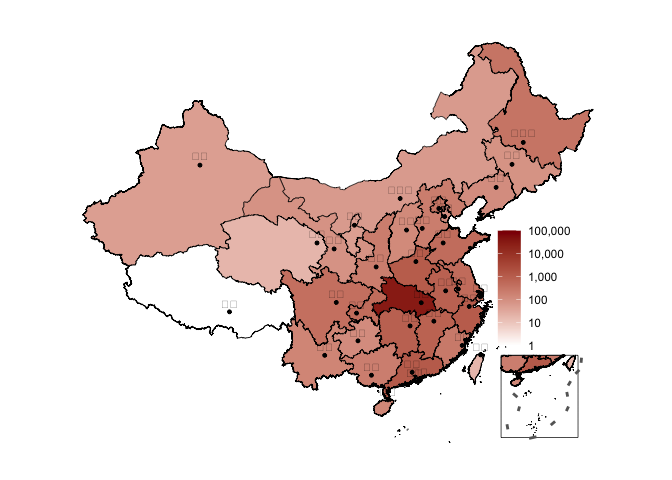
plot_map(ncov$area)
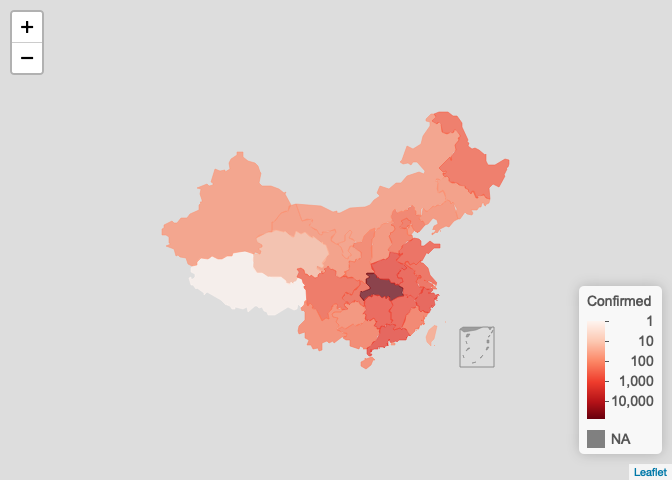
# log scale plot_map(ncov$area, scale = "log")
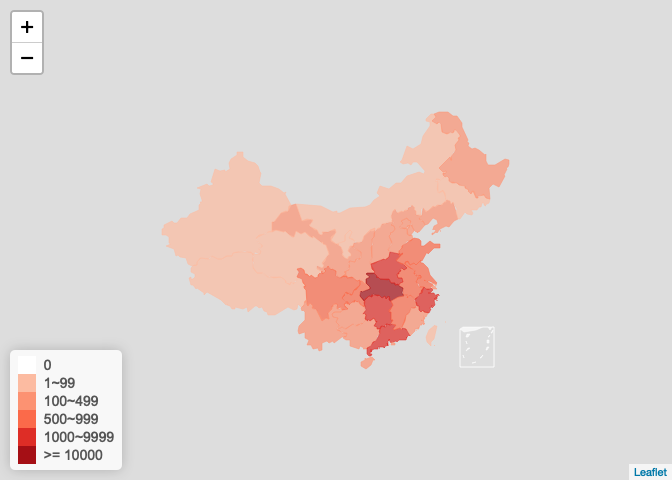
更进一步使用plot_china_map()可通过设置bins参数控制如何分组以填充不同的颜色, 自动把ncov为0的地区(包括南海驻岛)填充为白色
plot_china_map( china_ncov, bins = c(1, 100, 500, 1000, 10000), legend_position = "bottomleft" )
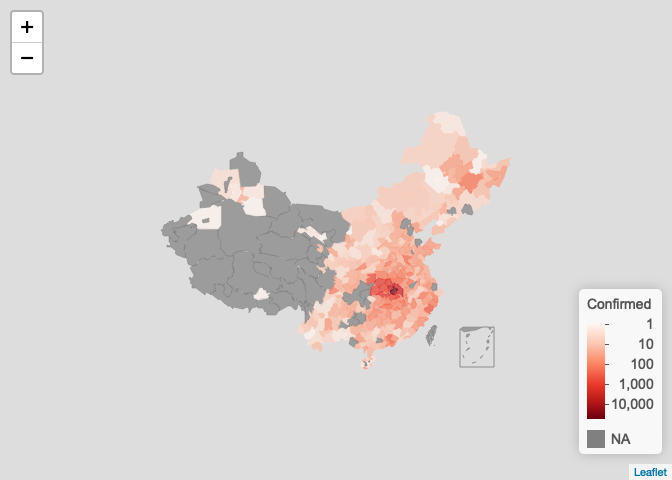
按城市显示
plot_map(ncov$area, method = "city", scale = "log")
ggplot
library(ggplot2) ncov$area$date <- as.Date(ncovr:::conv_time(ncov$area$updateTime)) choose_date <- "2020-02-10" x <- ncov$area[ncov$area$date <= as.Date(choose_date), ] x <- x[!duplicated(x$provinceName), ] plot_ggmap(x)
## Parsed with column specification:
## cols(
## province = col_character(),
## city = col_character(),
## long = col_double(),
## lat = col_double()
## )
省疫情图
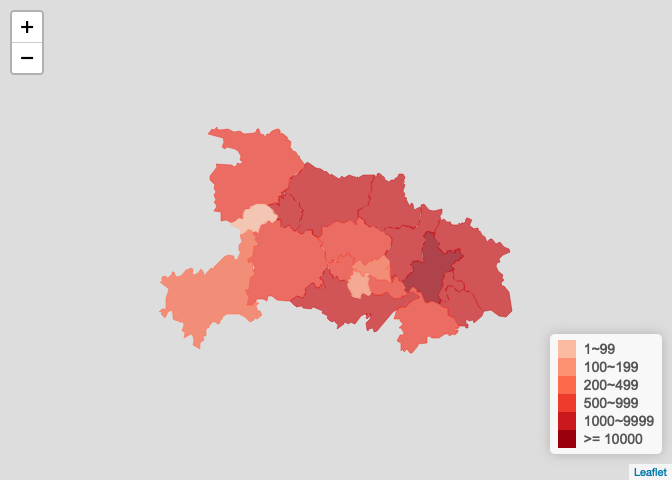
湖北省疫情图
# plot_province_map(ncov2, "<U+6E56><U+5317><U+7701>"),<U+6216> plot_province_map(hubei_ncov, "<U+6E56><U+5317><U+7701>", bins = c(1, 100, 200, 500, 1000, 10000))
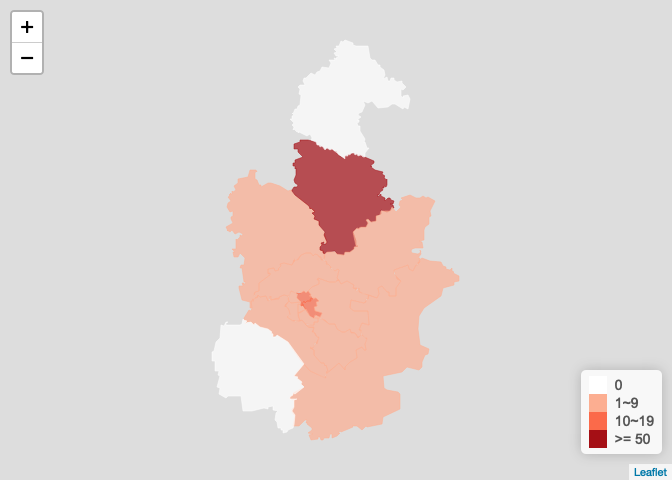
或直接基于ncov作图,无需提前取各省ncov数据,天津疫情图
plot_province_map(ncov2, "<U+5929><U+6D25><U+5E02>", bins = c(1, 10, 20, 50))
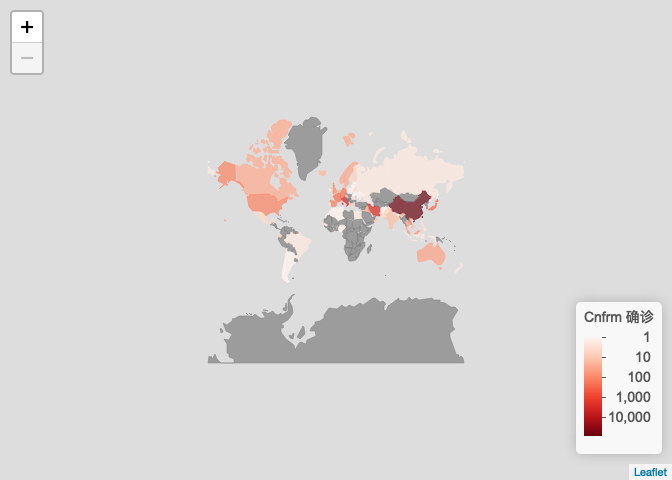
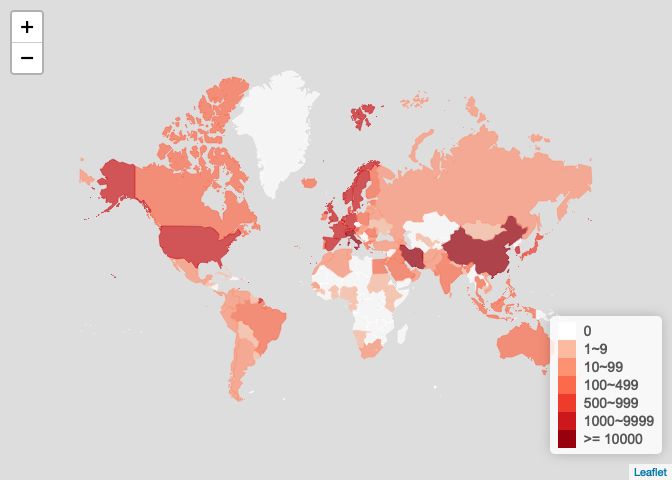
世界地图:各国疫情图
ncov$area$date <- as.Date(ncovr:::conv_time(ncov$area$updateTime)) ncov$area <- ncov$area[rev(order(ncov$area$date)), ] ncov_date <- as.character(Sys.Date()) y <- ncov$area[ncov$area$date <= as.Date(ncov_date), ] y <- y[!duplicated(y$provinceName), ] countryname <- data.frame( ncovr = c("United Kiongdom", "United States of America", "New Zealand", "Kampuchea (Cambodia )" ), leafletNC = c("UnitedKingdom", "UnitedStates", "NewZealand", "Cambodia" ), stringsAsFactors = FALSE ) x <- data.frame( countryEnglishName = y$countryEnglishName, countryName = y$countryName, confirmedCount = y$confirmedCount, stringsAsFactors = FALSE ) loc <- which(x$countryEnglishName %in% countryname$ncovr) x$countryEnglishName[loc] <- countryname$leafletNC[ match(x$countryEnglishName[loc], countryname$ncovr) ] x$countryEnglishName2 = x$countryEnglishName # for taiwan x_other <- x[!is.na(x$countryEnglishName) & x$countryEnglishName != 'China', ] x_china <- data.frame( countryEnglishName = 'China', countryName = unique(x[!is.na(x$countryEnglishName) & x$countryEnglishName == 'China', 'countryName']), confirmedCount = sum(x[!is.na(x$countryEnglishName) & x$countryEnglishName == 'China', 'confirmedCount']), countryEnglishName2 = 'China' ) x_taiwan <- x_china x_taiwan$countryEnglishName2 = "Taiwan" x <- rbind(x_other, x_china, x_taiwan) plot_map( x = x, key = "confirmedCount", scale = "log", method = 'country', legend_title = paste0("Cnfrm <U+786E><U+8BCA>"), filter = '<U+5F85><U+660E><U+786E><U+5730><U+533A>' )
更进一步
plot_world_map(world_ncov)
## Parsed with column specification:
## cols(
## name = col_character(),
## name_zh = col_character()
## )
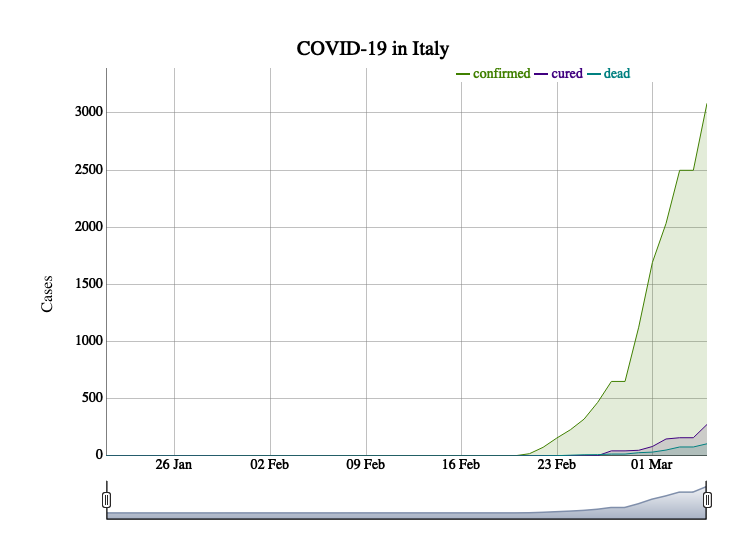
ts
x_ts <- ncov$area[, c('countryEnglishName', 'countryName', 'date', 'confirmedCount', 'curedCount', 'deadCount')] %>% dplyr::group_by(countryEnglishName, date) %>% dplyr::summarise( confirmed = max(confirmedCount), cured = max(curedCount), dead = max(deadCount)) %>% dplyr::ungroup() %>% dplyr::filter(!is.na(countryEnglishName) & !countryEnglishName == 'China') %>% as.data.frame() loc <- which(x_ts$countryEnglishName %in% countryname$ncovr) x_ts$countryEnglishName[loc] <- countryname$leafletNC[ match(x_ts$countryEnglishName[loc], countryname$ncovr) ] plot_ts( x_ts, area = "Italy", area_col = "countryEnglishName", date_col = "date", ts_col = c("confirmed", "cured", "dead") )
国外疫情图
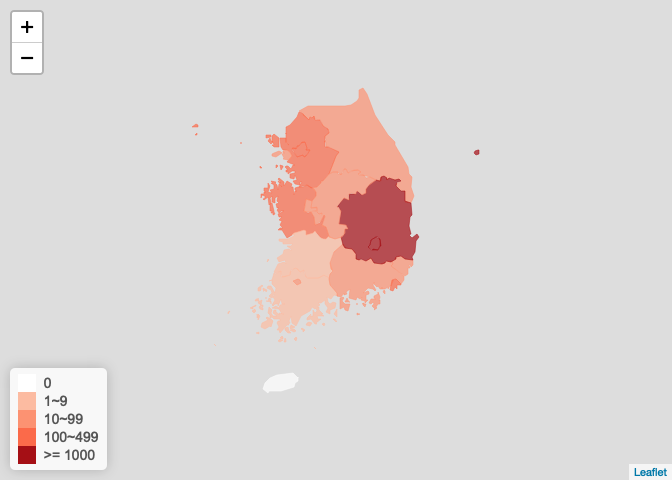
韩国疫情图
korea_ncov <- get_foreign_ncov("<U+97E9><U+56FD>") plot_foreign_map(korea_ncov, "korea")
## Parsed with column specification:
## cols(
## name = col_character(),
## name_zh = col_character(),
## provinceName = col_character(),
## provinceEnglishName = col_character()
## )
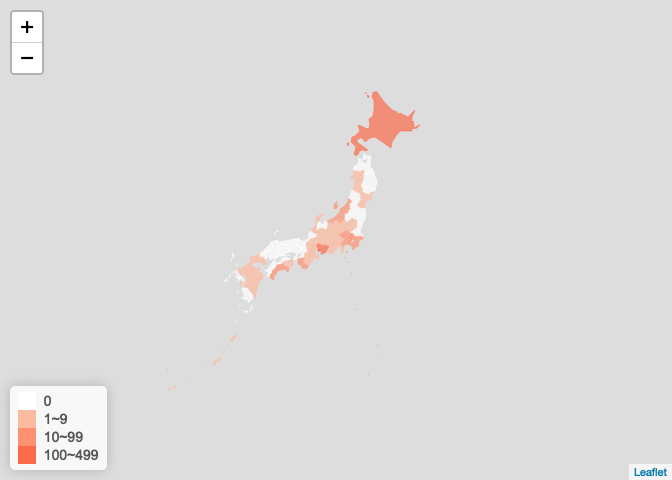
日本疫情图
jp_ncov <- get_foreign_ncov("<U+65E5><U+672C>") plot_foreign_map(jp_ncov, "japan")
## Parsed with column specification:
## cols(
## name = col_character(),
## name_zh = col_character(),
## provinceName = col_character(),
## provinceEnglishName = col_character()
## )
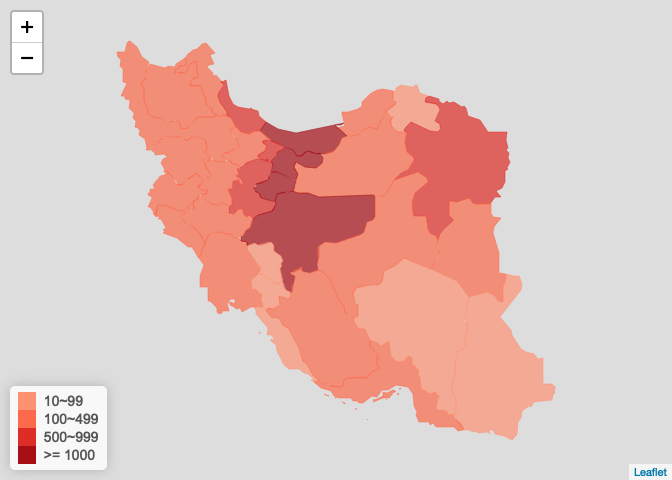
伊朗疫情图
iran_ncov <- get_foreign_ncov("<U+4F0A><U+6717>") plot_foreign_map(iran_ncov, "iran")
## Parsed with column specification:
## cols(
## name = col_character(),
## name_zh = col_character(),
## provinceName = col_character(),
## provinceEnglishName = col_character()
## )
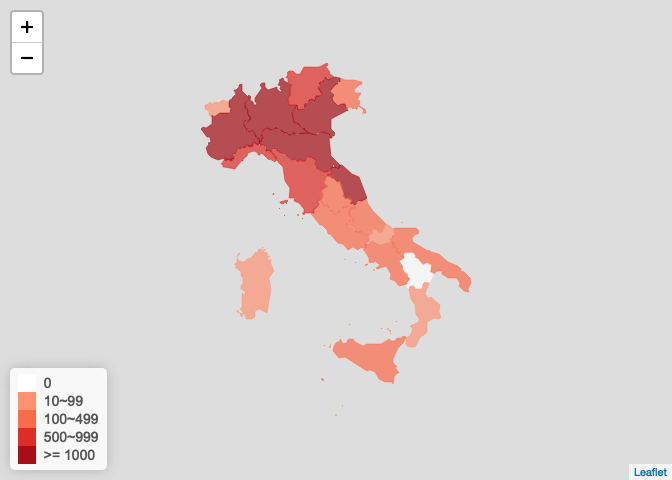
意大利疫情图
italy_ncov <- get_foreign_ncov("<U+610F><U+5927><U+5229>") plot_foreign_map(italy_ncov, "italy")
## Parsed with column specification:
## cols(
## name = col_character(),
## name_zh = col_character(),
## provinceName = col_character(),
## provinceEnglishName = col_character()
## )
直接画这四个国家的疫情图
foreign_countries <- c("<U+97E9><U+56FD>", "<U+4F0A><U+6717>", "<U+65E5><U+672C>", "<U+610F><U+5927><U+5229>") names(foreign_countries) <- c("korea", "iran", "japan", "italy") htmltools::tagList(purrr::imap( foreign_countries, ~ get_foreign_ncov(.x) %>% plot_foreign_map(.y) ))
更多功能请参看函数的帮助信息